Release Date
January 18, 2021
Study Abbreviations
| Full Name | Abbreviation |
|---|---|
| Amsterdam UMC COVID study group | Amsterdam_UMC_COVID_study_group |
| Ancestry | Ancestry |
| Genetic determinants of COVID-19 complications in the Brazilian population | BRACOVID |
| Genetic modifiers for COVID-19 related illness | BelCovid |
| Biobanque Quebec COVID19 | BQC19 |
| deCODE | DECODE |
| (Genetics of COVID-related Manifestation) | Corea |
| Estonian Biobank | EstBB |
| FinnGen | FinnGen |
| GEN-COVID, reCOVID | GENCOVID |
| genomiCC | genomicc |
| Genes & Health | GNH |
| Generation Scotland | GS |
| COVID19-Host(a)ge | HOSTAGE |
| Helix Exome+ COVID-19 Phenotypes | Helix |
| UK Blood Donors Cohort | INTERVAL |
| Michigan Genomics Initiative | MGI |
| Million Veterans Program | MVP |
| Netherlands Twin Register | NTR |
| Partners Healthcare Biobank | PHBB |
| Penn Medicine Biobank | PMBB |
| Qatar Genome Program | QGP |
| Determining the Molecular Pathways and Genetic Predisposition of the Acute Inflammatory Process Caused by SARS-CoV-2 | SPGRX |
| Genomic epidemiology of SARS-Cov-2 and host genetics in Coronavirus Disease 2019 (COVID-19) | Stanford |
| The genetic predisposition to severe COVID-19 | SweCovid |
| UK Biobank | UKBB |
| Bonn Study of COVID19 genetics | BoSCO |
| 23andMe | 23ANDME |
| Italy COVID19-Host(a)ge | Italy_HOSTAGE |
| Spain COVID19-Host(a)ge | Spain_HOSTAGE |
| Genomics England | genomicsengland100kgp |
| Lifelines | Lifelines |
| Gene Risk | GeneRISK |
| Rotterdam Study | RS |
| Columbia University COVID19 Biobank | CU |
| Geisinger Health System | GHS_Freeze_145 |
| Japan Coronavirus Taskforce | JapanTaskForce |
| 24Genetics | idipaz24genetics |
| FHoGID | FHoGID |
| UCLA Precision Health COVID-19 Biobank | UCLA |
| Genetic influences on severity of COVID-19 illness in Korea | Genetics_COVID19_Korea |
| Latvia COVID-19 research platform | LGDB |
| ACCOuNT | ACCOuNT |
| Vanderbilt Biobank | BioVU |
| The Colorado Center for Personalized Medicine | CCPM |
| Genomes for Life | GCAT |
| Genes for Good | GFG |
| Genotek COVID-19 study | Genotek |
| Mount Sinai Health System COVID-19 Genomics Initiative | MSHS_CGI |
| CHRIS | TOPMed_CHRIS10K |
Data Columns
| Column Name | Description |
|---|---|
| #CHR | chromosome |
| POS | chromosome position |
| REF | reference and non-effect allele |
| ALT | alternative and effect allele (beta is for this allele) |
| SNP | #CHR:POS:REF:ALT |
| all_meta_N | number of studies that had the variant after AF and INFO filtering and as such were used for the meta |
| all_inv_var_meta_beta | effect size on log(OR) scale |
| all_inv_var_meta_sebeta | standard error of effect size |
| all_inv_var_meta_p | p-value |
| all_inv_var_het_p | p-value from Cochran's Q heterogeneity test |
| all_meta_sample_N | total sample size |
| all_meta_AF | allele frequency in the meta-analysis |
| rsid | risd |
Release Notes
Meta-analysis was done with fixed effects inverse variance weighting. Results are available in genome builds 38 and 37. An AF filter of 0.001 and an INFO filter of 0.6 was applied to each study before meta.
Extras
**New** the full downloads below do not contain the 23andMe samples (the most well-powered analyses), however the top 10K SNPs from analyses that include 23andMe samples are available here.
There are additional leave-one-out results not listed here available at this storage bucket: gs://covid19-hg-public/20201215/results/20210107. You will need gsutil to list and download these files.
Note: variants with heterogeneity p-value less than 0.001 across studies are in green.
A2_ALL_leave_23andme
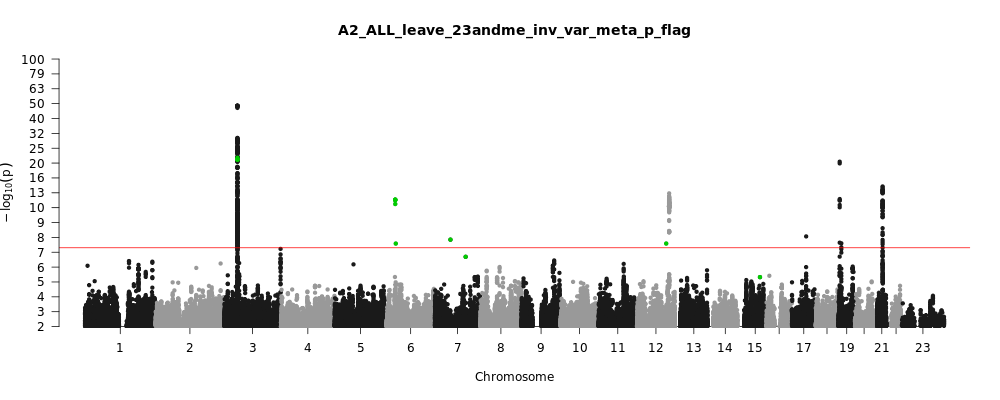
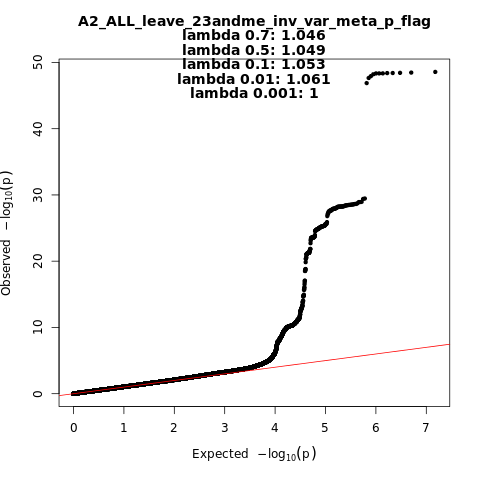
Phenotype
Very severe respiratory confirmed covid vs. population
Population
All
Total Cases
5582
Total Controls
709010
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 88 | 552 |
| BRACOVID_AMR | 539 | 1149 |
| BelCovid_EUR | 182 | 1477 |
| CU_AFR | 133 | 2610 |
| CU_EUR | 203 | 2149 |
| FinnGen_FIN | 68 | 238643 |
| GENCOVID_EUR | 724 | 2443 |
| GHS_Freeze_145_EUR | 53 | 112862 |
| JapanTaskForce_EAS | 155 | 1705 |
| SweCovid_EUR | 77 | 3748 |
| UKBB_EUR | 309 | 328577 |
| genomicc_EAS | 149 | 745 |
| idipaz24genetics_EUR | 59 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 66 | 1413 |
| SPGRX_EUR | 101 | 302 |
| genomicc_EUR | 1676 | 8380 |
| Italy_HOSTAGE_EUR | 698 | 1255 |
| Spain_HOSTAGE_EUR | 302 | 925 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_A2_ALL_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_A2_ALL_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_leave_23andme_20210107.txt.gz_1.0E-5.txt
A2_ALL_leave_UKBB
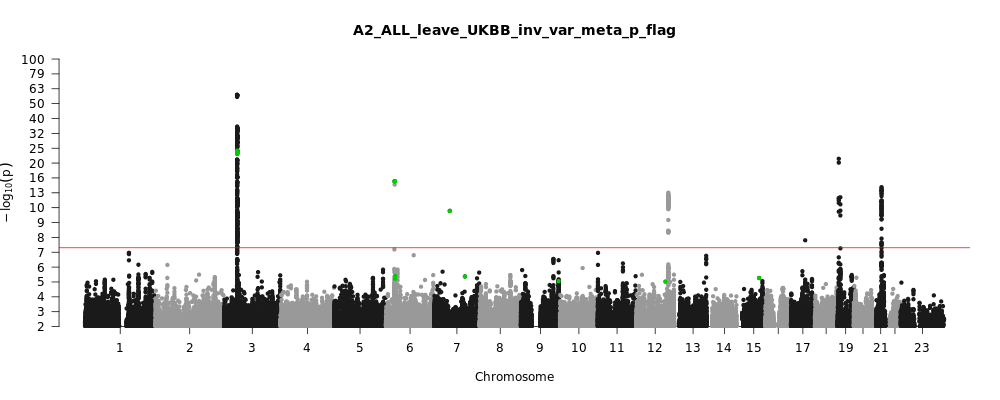
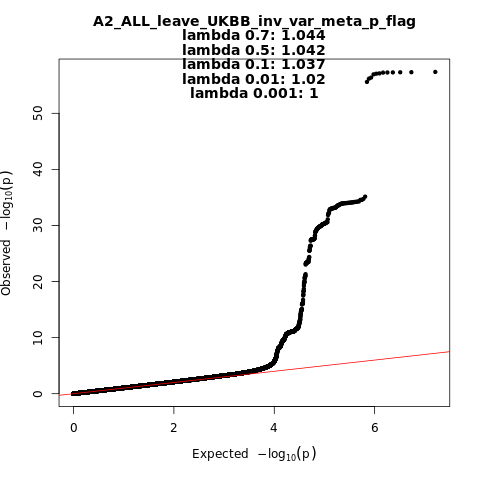
Phenotype
Very severe respiratory confirmed covid vs. population
Population
All
Total Cases
5870
Total Controls
1155203
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 88 | 552 |
| BRACOVID_AMR | 539 | 1149 |
| BelCovid_EUR | 182 | 1477 |
| CU_AFR | 133 | 2610 |
| CU_EUR | 203 | 2149 |
| FinnGen_FIN | 68 | 238643 |
| GENCOVID_EUR | 724 | 2443 |
| GHS_Freeze_145_EUR | 53 | 112862 |
| JapanTaskForce_EAS | 155 | 1705 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EAS | 149 | 745 |
| idipaz24genetics_EUR | 59 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 66 | 1413 |
| SPGRX_EUR | 101 | 302 |
| genomicc_EUR | 1676 | 8380 |
| Italy_HOSTAGE_EUR | 698 | 1255 |
| Spain_HOSTAGE_EUR | 302 | 925 |
| 23ANDME_EUR | 495 | 680440 |
| 23ANDME_HIS | 102 | 94330 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_leave_UKBB_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_leave_UKBB_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_A2_ALL_leave_UKBB_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_A2_ALL_leave_UKBB_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_leave_UKBB_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_leave_UKBB_23andme_20210107.txt.gz_1.0E-5.txt
A2_ALL_eur
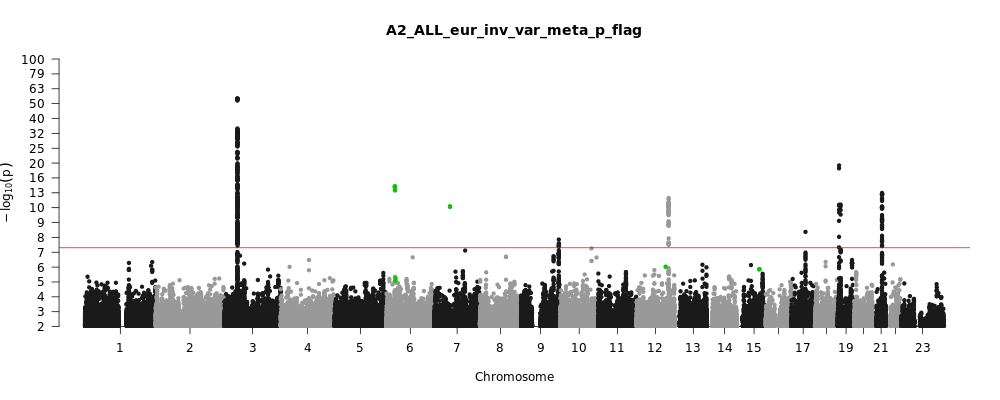
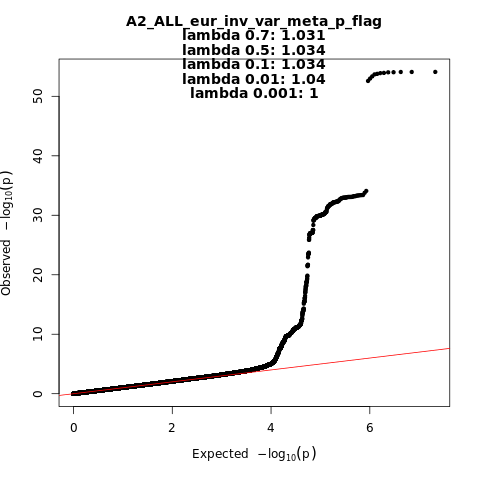
Phenotype
Very severe respiratory confirmed covid vs. population
Population
Eur
Total Cases
5101
Total Controls
1383241
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 88 | 552 |
| BelCovid_EUR | 182 | 1477 |
| CU_EUR | 203 | 2149 |
| FinnGen_FIN | 68 | 238643 |
| GENCOVID_EUR | 724 | 2443 |
| GHS_Freeze_145_EUR | 53 | 112862 |
| SweCovid_EUR | 77 | 3748 |
| UKBB_EUR | 309 | 328577 |
| idipaz24genetics_EUR | 59 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 66 | 1413 |
| SPGRX_EUR | 101 | 302 |
| genomicc_EUR | 1676 | 8380 |
| Italy_HOSTAGE_EUR | 698 | 1255 |
| Spain_HOSTAGE_EUR | 302 | 925 |
| 23ANDME_EUR | 495 | 680440 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_eur_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_eur_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_A2_ALL_eur_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_A2_ALL_eur_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_eur_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_eur_leave_23andme_20210107.txt.gz_1.0E-5.txt
A2_ALL_eur_leave_ukbb
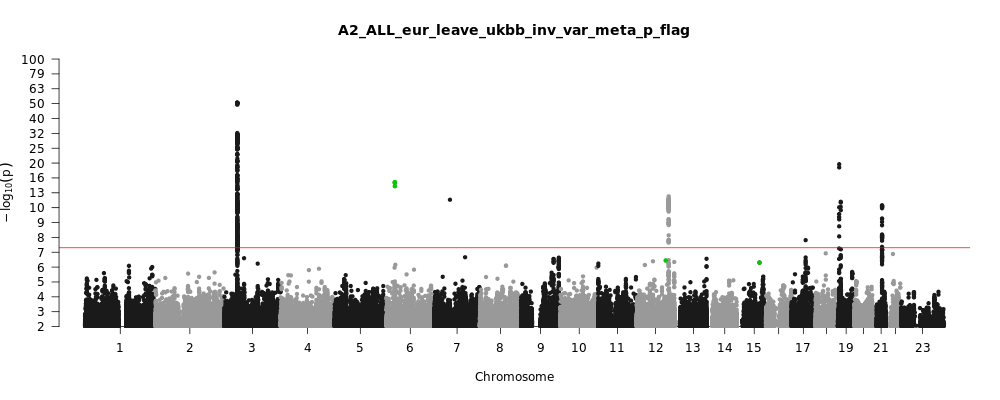
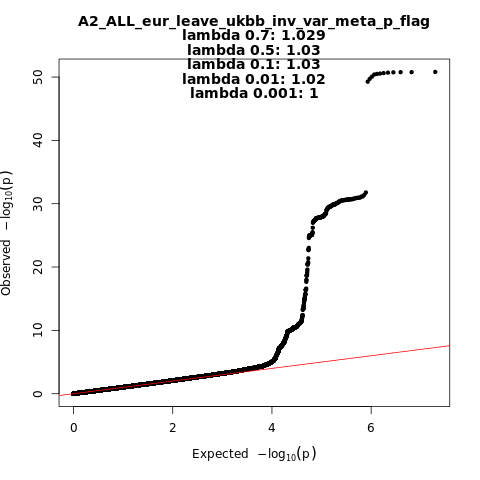
Phenotype
Very severe respiratory confirmed covid vs. population
Population
Eur
Total Cases
4792
Total Controls
1054664
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 88 | 552 |
| BelCovid_EUR | 182 | 1477 |
| CU_EUR | 203 | 2149 |
| FinnGen_FIN | 68 | 238643 |
| GENCOVID_EUR | 724 | 2443 |
| GHS_Freeze_145_EUR | 53 | 112862 |
| SweCovid_EUR | 77 | 3748 |
| idipaz24genetics_EUR | 59 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 66 | 1413 |
| SPGRX_EUR | 101 | 302 |
| genomicc_EUR | 1676 | 8380 |
| Italy_HOSTAGE_EUR | 698 | 1255 |
| Spain_HOSTAGE_EUR | 302 | 925 |
| 23ANDME_EUR | 495 | 680440 |
Downloads
GRCh37 liftover: COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_A2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz_1.0E-5.txt
B1_ALL_leave_23andme
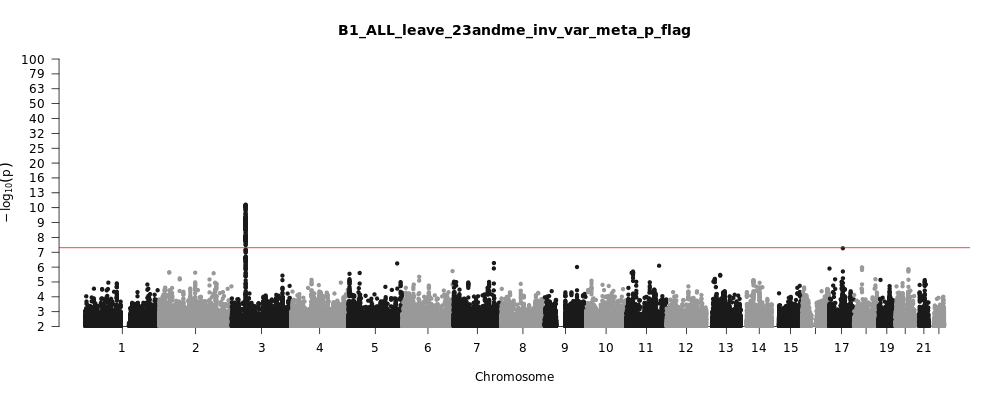
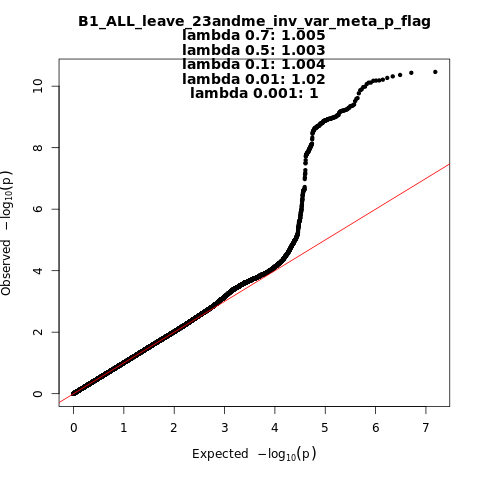
Phenotype
Hospitalized covid vs. not hospitalized covid
Population
All
Total Cases
5773
Total Controls
15497
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BelCovid_EUR | 361 | 122 |
| BoSCO_EUR | 212 | 512 |
| EstBB_EUR | 60 | 512 |
| FHoGID_EUR | 362 | 259 |
| FinnGen_FIN | 106 | 520 |
| GENCOVID_EUR | 892 | 249 |
| GNH_SAS | 115 | 1264 |
| UCLA_AMR | 95 | 74 |
| UCLA_EUR | 80 | 123 |
| UKBB_AFR | 71 | 119 |
| UKBB_EUR | 1670 | 4610 |
| UKBB_SAS | 71 | 230 |
| SPGRX_EUR | 311 | 51 |
| DECODE_EUR | 89 | 1808 |
| PMBB_AFR | 66 | 100 |
| QGP_ARAB | 60 | 640 |
| MVP_AFR | 349 | 862 |
| MVP_EUR | 436 | 1083 |
| MVP_HIS | 117 | 392 |
| Ancestry_EUR | 250 | 1967 |
Downloads
GRCh37 liftover: COVID19_HGI_B1_ALL_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B1_ALL_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B1_ALL_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B1_ALL_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B1_ALL_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B1_ALL_leave_23andme_20210107.txt.gz_1.0E-5.txt
B1_ALL_leave_UKBB
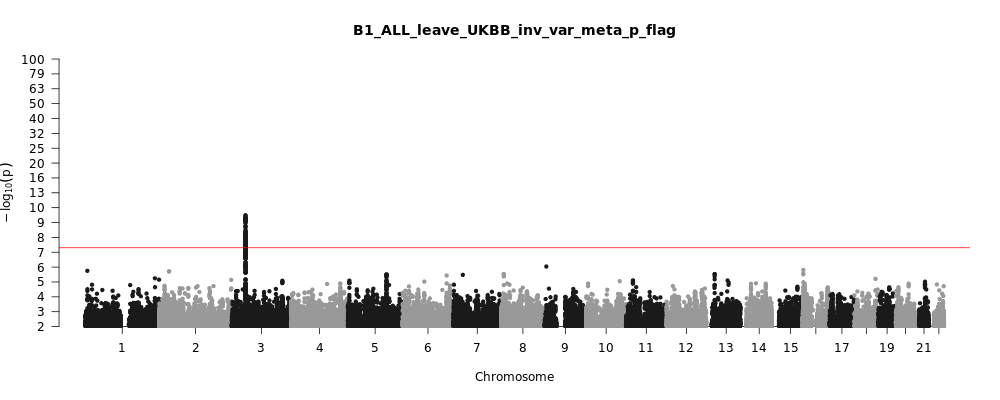
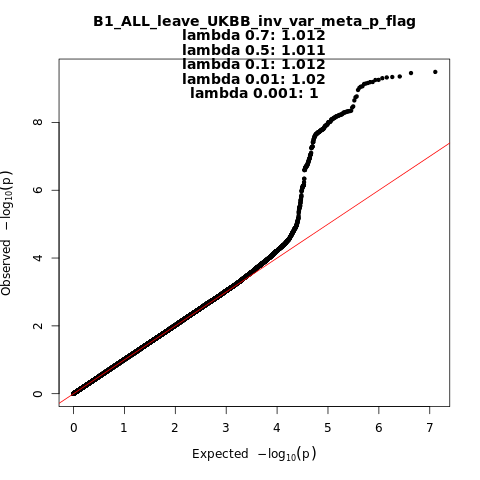
Phenotype
Hospitalized covid vs. not hospitalized covid
Population
All
Total Cases
3961
Total Controls
10538
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BelCovid_EUR | 361 | 122 |
| BoSCO_EUR | 212 | 512 |
| EstBB_EUR | 60 | 512 |
| FHoGID_EUR | 362 | 259 |
| FinnGen_FIN | 106 | 520 |
| GENCOVID_EUR | 892 | 249 |
| GNH_SAS | 115 | 1264 |
| UCLA_AMR | 95 | 74 |
| UCLA_EUR | 80 | 123 |
| SPGRX_EUR | 311 | 51 |
| DECODE_EUR | 89 | 1808 |
| PMBB_AFR | 66 | 100 |
| QGP_ARAB | 60 | 640 |
| MVP_AFR | 349 | 862 |
| MVP_EUR | 436 | 1083 |
| MVP_HIS | 117 | 392 |
| Ancestry_EUR | 250 | 1967 |
Downloads
GRCh37 liftover: COVID19_HGI_B1_ALL_leave_UKBB_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B1_ALL_leave_UKBB_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B1_ALL_leave_UKBB_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B1_ALL_leave_UKBB_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B1_ALL_leave_UKBB_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B1_ALL_leave_UKBB_23andme_20210107.txt.gz_1.0E-5.txt
B1_ALL_eur
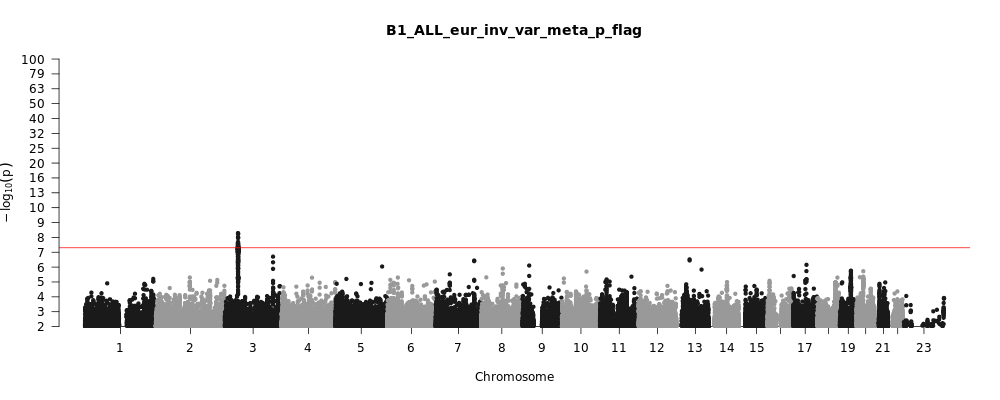
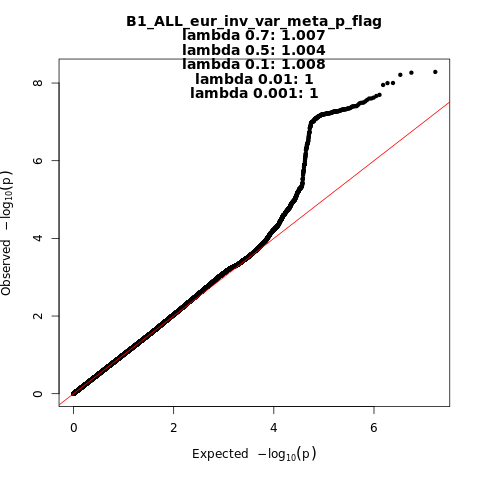
Phenotype
Hospitalized covid vs. not hospitalized covid
Population
Eur
Total Cases
4829
Total Controls
11816
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BelCovid_EUR | 361 | 122 |
| BoSCO_EUR | 212 | 512 |
| EstBB_EUR | 60 | 512 |
| FHoGID_EUR | 362 | 259 |
| FinnGen_FIN | 106 | 520 |
| GENCOVID_EUR | 892 | 249 |
| UCLA_EUR | 80 | 123 |
| UKBB_EUR | 1670 | 4610 |
| SPGRX_EUR | 311 | 51 |
| DECODE_EUR | 89 | 1808 |
| MVP_EUR | 436 | 1083 |
| Ancestry_EUR | 250 | 1967 |
Downloads
GRCh37 liftover: COVID19_HGI_B1_ALL_eur_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B1_ALL_eur_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B1_ALL_eur_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B1_ALL_eur_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B1_ALL_eur_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B1_ALL_eur_leave_23andme_20210107.txt.gz_1.0E-5.txt
B1_ALL_eur_leave_ukbb
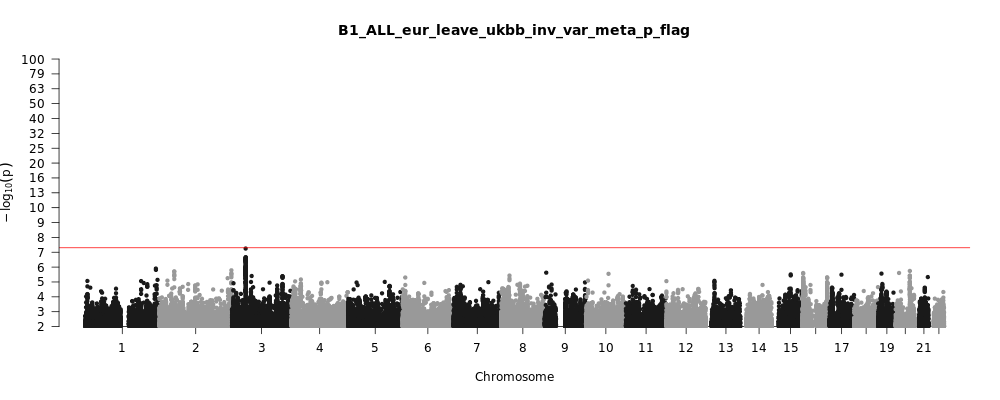
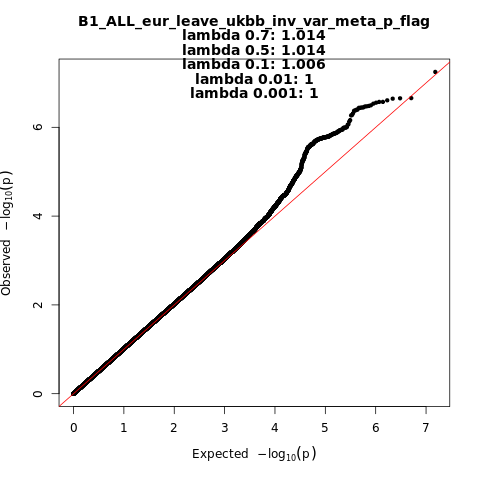
Phenotype
Hospitalized covid vs. not hospitalized covid
Population
Eur
Total Cases
3159
Total Controls
7206
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BelCovid_EUR | 361 | 122 |
| BoSCO_EUR | 212 | 512 |
| EstBB_EUR | 60 | 512 |
| FHoGID_EUR | 362 | 259 |
| FinnGen_FIN | 106 | 520 |
| GENCOVID_EUR | 892 | 249 |
| UCLA_EUR | 80 | 123 |
| SPGRX_EUR | 311 | 51 |
| DECODE_EUR | 89 | 1808 |
| MVP_EUR | 436 | 1083 |
| Ancestry_EUR | 250 | 1967 |
Downloads
GRCh37 liftover: COVID19_HGI_B1_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B1_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B1_ALL_eur_leave_ukbb_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B1_ALL_eur_leave_ukbb_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B1_ALL_eur_leave_ukbb_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B1_ALL_eur_leave_ukbb_23andme_20210107.txt.gz_1.0E-5.txt
B2_ALL_leave_23andme
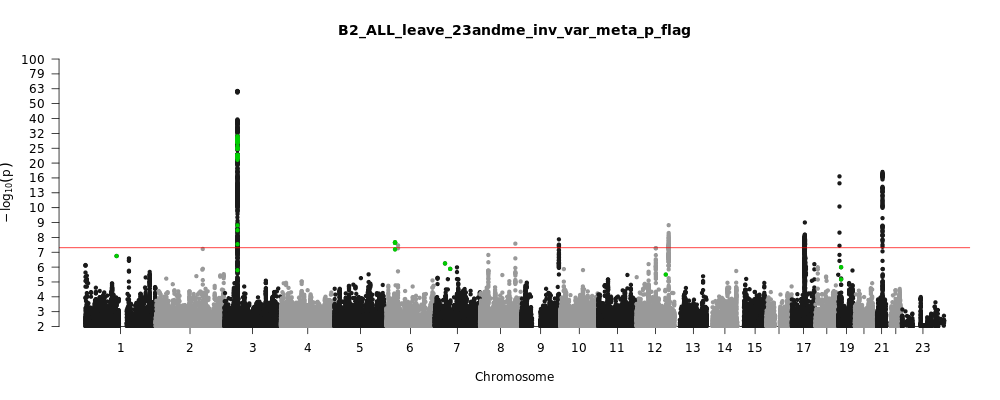
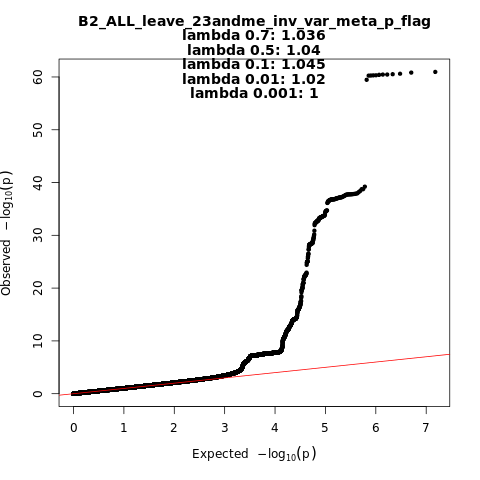
Phenotype
Hospitalized covid vs. population
Population
All
Total Cases
12888
Total Controls
1295966
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 244 | 396 |
| BRACOVID_AMR | 853 | 835 |
| BelCovid_EUR | 363 | 1477 |
| CU_AFR | 304 | 2610 |
| CU_EUR | 453 | 2149 |
| EstBB_EUR | 90 | 196339 |
| FinnGen_FIN | 106 | 238605 |
| GENCOVID_EUR | 893 | 2443 |
| GHS_Freeze_145_EUR | 180 | 112862 |
| GNH_SAS | 115 | 34049 |
| Genetics_COVID19_Korea_EAS | 624 | 6549 |
| JapanTaskForce_EAS | 572 | 1705 |
| LGDB_EUR | 57 | 1531 |
| UCLA_AMR | 95 | 4569 |
| UCLA_EUR | 80 | 17514 |
| UKBB_AFR | 71 | 7691 |
| UKBB_EUR | 1670 | 328577 |
| UKBB_SAS | 71 | 9231 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| SPGRX_EUR | 311 | 302 |
| DECODE_EUR | 89 | 274322 |
| PMBB_AFR | 66 | 8536 |
| QGP_ARAB | 60 | 13360 |
| MVP_AFR | 349 | 1745 |
| MVP_EUR | 436 | 2180 |
| MVP_HIS | 117 | 585 |
| Corea_EAS | 69 | 6500 |
| HOSTAGE_EUR | 1610 | 2205 |
| BoSCO_EUR | 212 | 512 |
| FHoGID_EUR | 362 | 259 |
| Ancestry_EUR | 250 | 1967 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EAS | 149 | 745 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B2_ALL_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B2_ALL_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_leave_23andme_20210107.txt.gz_1.0E-5.txt
B2_ALL_leave_UKBB
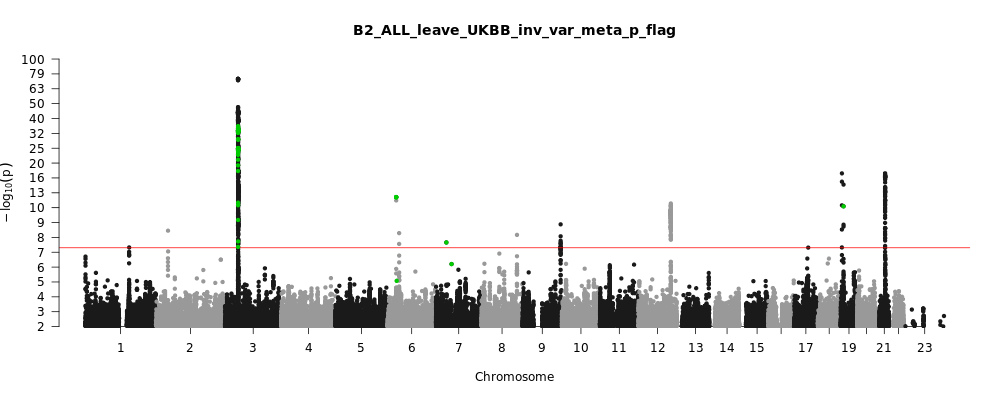
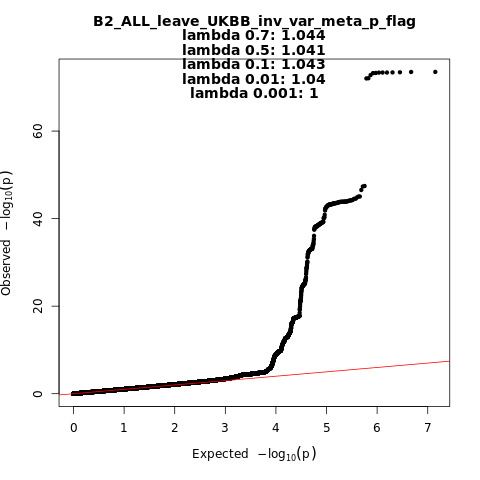
Phenotype
Hospitalized covid vs. population
Population
All
Total Cases
11829
Total Controls
1725210
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 244 | 396 |
| BRACOVID_AMR | 853 | 835 |
| BelCovid_EUR | 363 | 1477 |
| CU_AFR | 304 | 2610 |
| CU_EUR | 453 | 2149 |
| EstBB_EUR | 90 | 196339 |
| FinnGen_FIN | 106 | 238605 |
| GENCOVID_EUR | 893 | 2443 |
| GHS_Freeze_145_EUR | 180 | 112862 |
| GNH_SAS | 115 | 34049 |
| Genetics_COVID19_Korea_EAS | 624 | 6549 |
| JapanTaskForce_EAS | 572 | 1705 |
| LGDB_EUR | 57 | 1531 |
| UCLA_AMR | 95 | 4569 |
| UCLA_EUR | 80 | 17514 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| SPGRX_EUR | 311 | 302 |
| DECODE_EUR | 89 | 274322 |
| PMBB_AFR | 66 | 8536 |
| QGP_ARAB | 60 | 13360 |
| MVP_AFR | 349 | 1745 |
| MVP_EUR | 436 | 2180 |
| MVP_HIS | 117 | 585 |
| Corea_EAS | 69 | 6500 |
| HOSTAGE_EUR | 1610 | 2205 |
| 23ANDME_EUR | 613 | 680416 |
| 23ANDME_HIS | 140 | 94327 |
| BoSCO_EUR | 212 | 512 |
| FHoGID_EUR | 362 | 259 |
| Ancestry_EUR | 250 | 1967 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EAS | 149 | 745 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_leave_UKBB_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_leave_UKBB_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B2_ALL_leave_UKBB_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B2_ALL_leave_UKBB_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_leave_UKBB_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_leave_UKBB_23andme_20210107.txt.gz_1.0E-5.txt
B2_ALL_eur
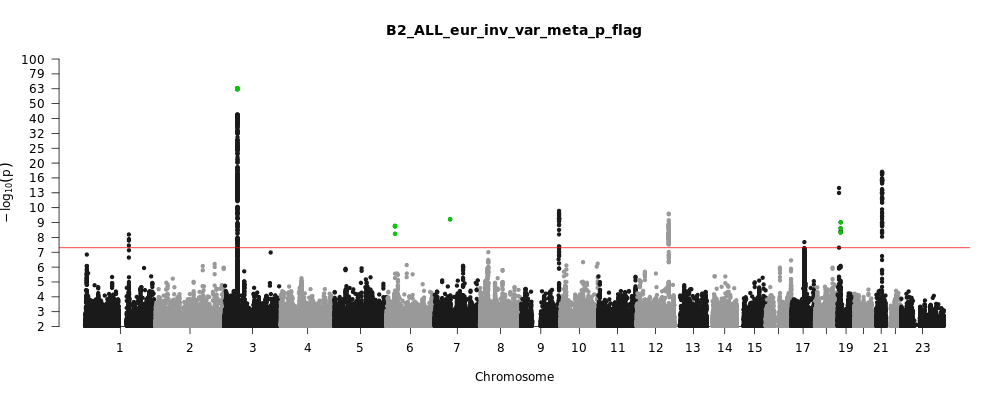
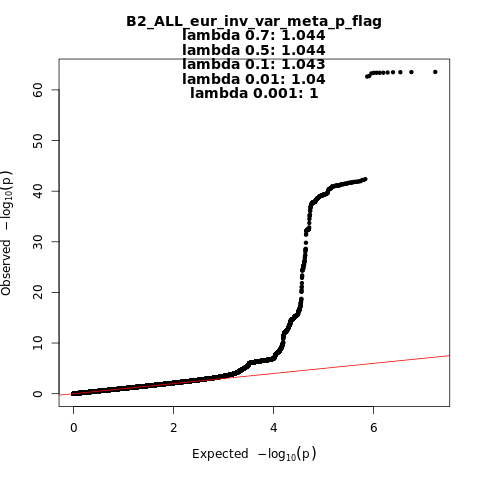
Phenotype
Hospitalized covid vs. population
Population
Eur
Total Cases
9986
Total Controls
1877672
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 244 | 396 |
| BelCovid_EUR | 363 | 1477 |
| CU_EUR | 453 | 2149 |
| EstBB_EUR | 90 | 196339 |
| FinnGen_FIN | 106 | 238605 |
| GENCOVID_EUR | 893 | 2443 |
| GHS_Freeze_145_EUR | 180 | 112862 |
| LGDB_EUR | 57 | 1531 |
| UCLA_EUR | 80 | 17514 |
| UKBB_EUR | 1670 | 328577 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| SPGRX_EUR | 311 | 302 |
| DECODE_EUR | 89 | 274322 |
| MVP_EUR | 436 | 2180 |
| HOSTAGE_EUR | 1610 | 2205 |
| 23ANDME_EUR | 613 | 680416 |
| BoSCO_EUR | 212 | 512 |
| FHoGID_EUR | 362 | 259 |
| Ancestry_EUR | 250 | 1967 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_eur_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_eur_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B2_ALL_eur_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B2_ALL_eur_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_eur_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_eur_leave_23andme_20210107.txt.gz_1.0E-5.txt
B2_ALL_eur_leave_ukbb
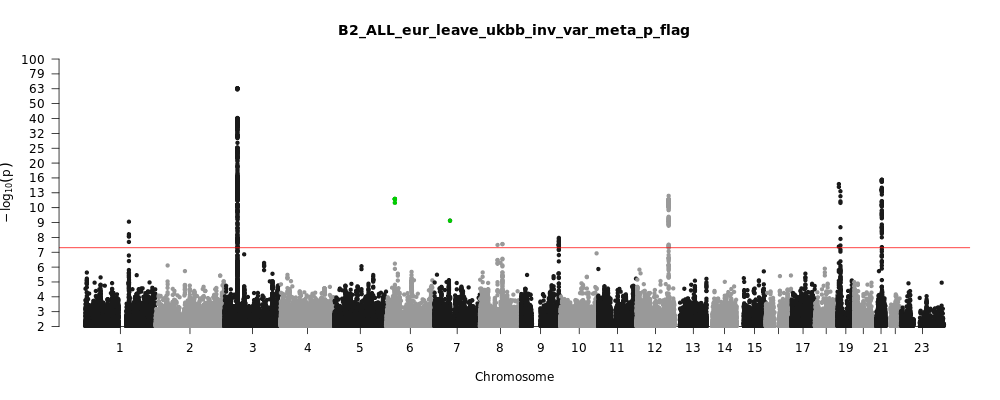
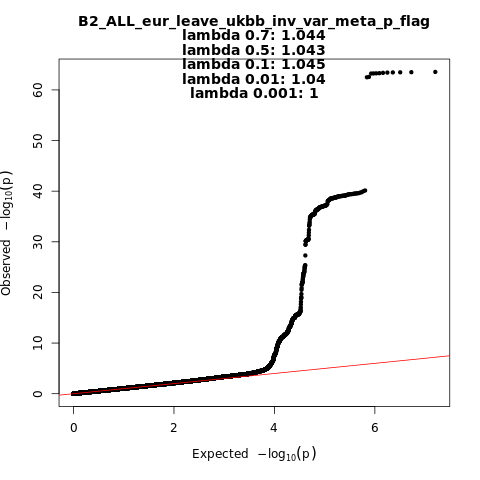
Phenotype
Hospitalized covid vs. population
Population
Eur
Total Cases
8316
Total Controls
1549095
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 244 | 396 |
| BelCovid_EUR | 363 | 1477 |
| CU_EUR | 453 | 2149 |
| EstBB_EUR | 90 | 196339 |
| FinnGen_FIN | 106 | 238605 |
| GENCOVID_EUR | 893 | 2443 |
| GHS_Freeze_145_EUR | 180 | 112862 |
| LGDB_EUR | 57 | 1531 |
| UCLA_EUR | 80 | 17514 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| SPGRX_EUR | 311 | 302 |
| DECODE_EUR | 89 | 274322 |
| MVP_EUR | 436 | 2180 |
| HOSTAGE_EUR | 1610 | 2205 |
| 23ANDME_EUR | 613 | 680416 |
| BoSCO_EUR | 212 | 512 |
| FHoGID_EUR | 362 | 259 |
| Ancestry_EUR | 250 | 1967 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_B2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz_1.0E-5.txt
C2_ALL_leave_23andme
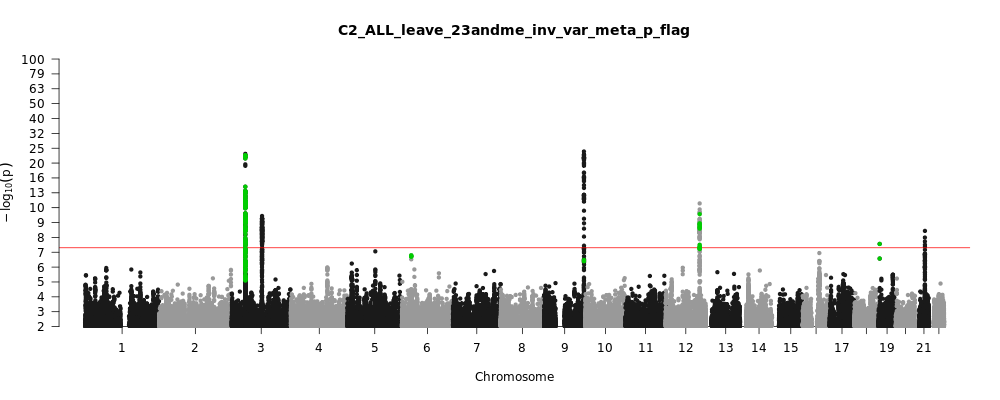
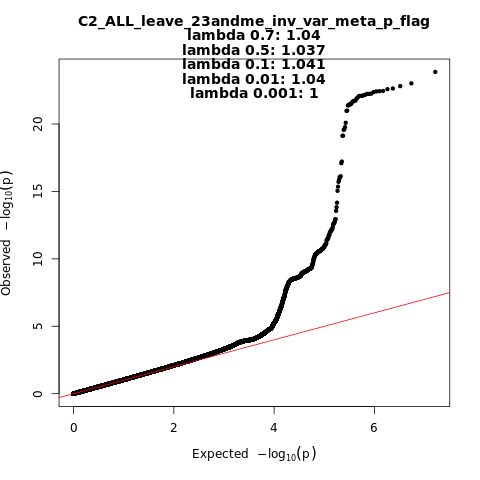
Phenotype
Covid vs. population
Population
All
Total Cases
36590
Total Controls
1668938
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ACCOuNT_AFR | 57 | 799 |
| BQC19_EUR | 269 | 368 |
| BelCovid_EUR | 485 | 1477 |
| BioVU_EUR | 141 | 70615 |
| CCPM_EUR | 332 | 32375 |
| CU_AFR | 332 | 2610 |
| CU_EUR | 508 | 2149 |
| DECODE_EUR | 4256 | 270934 |
| EstBB_EUR | 1322 | 193774 |
| FinnGen_FIN | 810 | 237901 |
| GCAT_EUR | 253 | 4735 |
| GENCOVID_EUR | 1220 | 2443 |
| GFG_EUR | 147 | 5442 |
| GHS_Freeze_145_EUR | 869 | 112862 |
| GNH_SAS | 1379 | 32785 |
| Genotek_EUR | 676 | 12317 |
| INTERVAL_EUR | 838 | 40994 |
| JapanTaskForce_EAS | 614 | 1705 |
| LGDB_EUR | 275 | 1313 |
| Lifelines_EUR | 244 | 26553 |
| MSHS_CGI_EUR | 330 | 1396 |
| Stanford_EUR | 169 | 190 |
| TOPMed_CHRIS10K_EUR | 92 | 2373 |
| TOPMed_Gardena_EUR | 452 | 458 |
| UCLA_AMR | 169 | 4495 |
| UCLA_EUR | 203 | 17391 |
| UKBB_AFR | 206 | 7691 |
| UKBB_EUR | 6490 | 328577 |
| UKBB_SAS | 309 | 9231 |
| SPGRX_EUR | 362 | 302 |
| PMBB_AFR | 166 | 8436 |
| QGP_ARAB | 700 | 13360 |
| MVP_AFR | 1217 | 6085 |
| MVP_EUR | 1520 | 7600 |
| MVP_HIS | 510 | 2550 |
| Corea_EAS | 108 | 6500 |
| genomicsengland100kgp_EUR | 218 | 62302 |
| Helix_EUR | 178 | 5441 |
| MGI_EUR | 122 | 51458 |
| NTR_EUR | 145 | 5252 |
| PHBB_AFR | 60 | 2445 |
| PHBB_EUR | 151 | 29966 |
| PHBB_HIS | 66 | 2405 |
| Ancestry_EUR | 2417 | 14933 |
| BRACOVID_AMR | 853 | 835 |
| Genetics_COVID19_Korea_EAS | 624 | 6549 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| HOSTAGE_EUR | 1610 | 2205 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EAS | 149 | 745 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_C2_ALL_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_C2_ALL_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_leave_23andme_20210107.txt.gz_1.0E-5.txt
C2_ALL_leave_UKBB
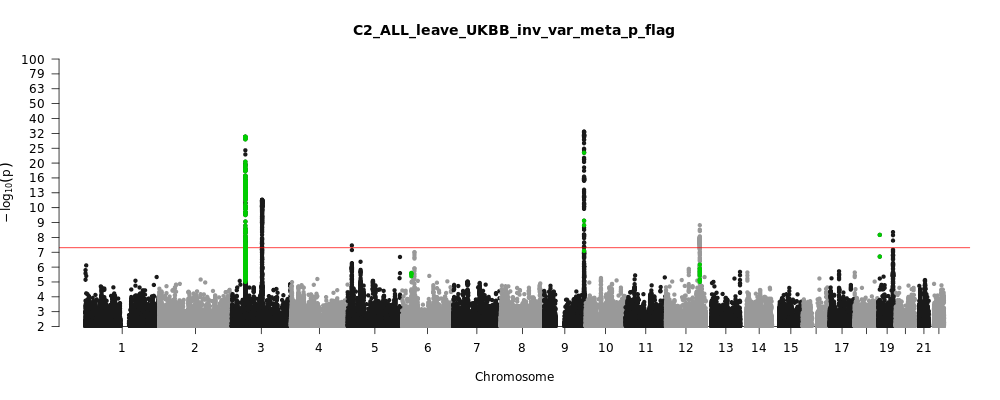
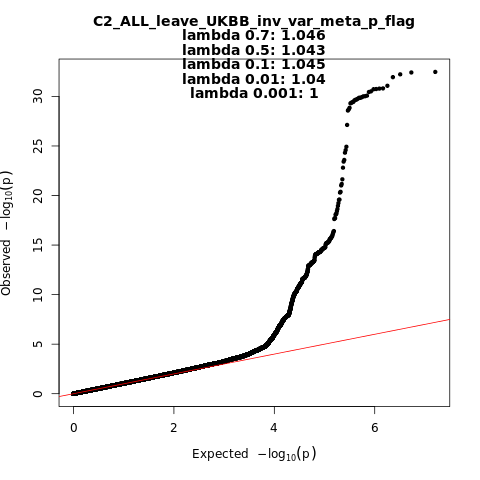
Phenotype
Covid vs. population
Population
All
Total Cases
42557
Total Controls
1424707
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| ACCOuNT_AFR | 57 | 799 |
| BQC19_EUR | 269 | 368 |
| BelCovid_EUR | 485 | 1477 |
| BioVU_EUR | 141 | 70615 |
| CCPM_EUR | 332 | 32375 |
| CU_AFR | 332 | 2610 |
| CU_EUR | 508 | 2149 |
| DECODE_EUR | 4256 | 270934 |
| EstBB_EUR | 1322 | 193774 |
| FinnGen_FIN | 810 | 237901 |
| GCAT_EUR | 253 | 4735 |
| GENCOVID_EUR | 1220 | 2443 |
| GFG_EUR | 147 | 5442 |
| GHS_Freeze_145_EUR | 869 | 112862 |
| GNH_SAS | 1379 | 32785 |
| Genotek_EUR | 676 | 12317 |
| INTERVAL_EUR | 838 | 40994 |
| JapanTaskForce_EAS | 614 | 1705 |
| LGDB_EUR | 275 | 1313 |
| Lifelines_EUR | 244 | 26553 |
| MSHS_CGI_EUR | 330 | 1396 |
| Stanford_EUR | 169 | 190 |
| TOPMed_CHRIS10K_EUR | 92 | 2373 |
| TOPMed_Gardena_EUR | 452 | 458 |
| UCLA_AMR | 169 | 4495 |
| UCLA_EUR | 203 | 17391 |
| SPGRX_EUR | 362 | 302 |
| PMBB_AFR | 166 | 8436 |
| QGP_ARAB | 700 | 13360 |
| MVP_AFR | 1217 | 6085 |
| MVP_EUR | 1520 | 7600 |
| MVP_HIS | 510 | 2550 |
| Corea_EAS | 108 | 6500 |
| genomicsengland100kgp_EUR | 218 | 62302 |
| Helix_EUR | 178 | 5441 |
| MGI_EUR | 122 | 51458 |
| NTR_EUR | 145 | 5252 |
| PHBB_AFR | 60 | 2445 |
| PHBB_EUR | 151 | 29966 |
| PHBB_HIS | 66 | 2405 |
| Ancestry_EUR | 2417 | 14933 |
| 23ANDME_AFR | 506 | 3110 |
| 23ANDME_EUR | 9913 | 85072 |
| 23ANDME_HIS | 2553 | 13086 |
| BRACOVID_AMR | 853 | 835 |
| Genetics_COVID19_Korea_EAS | 624 | 6549 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| HOSTAGE_EUR | 1610 | 2205 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EAS | 149 | 745 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_leave_UKBB_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_leave_UKBB_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_C2_ALL_leave_UKBB_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_C2_ALL_leave_UKBB_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_leave_UKBB_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_leave_UKBB_23andme_20210107.txt.gz_1.0E-5.txt
C2_ALL_eur
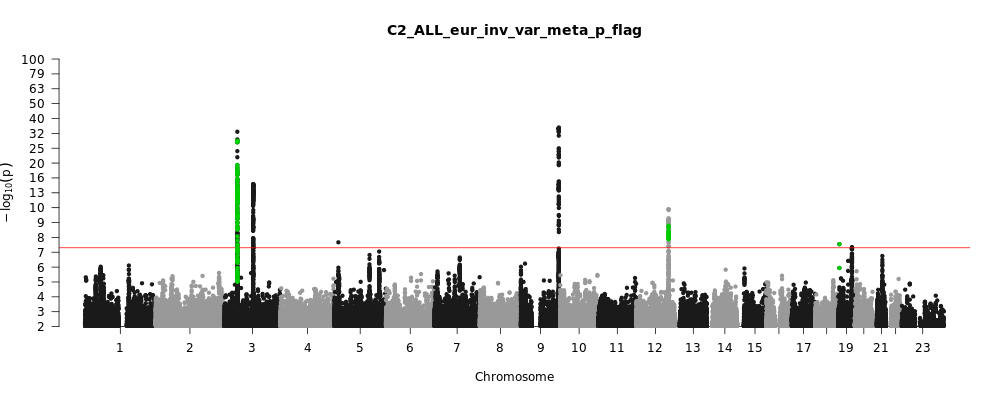
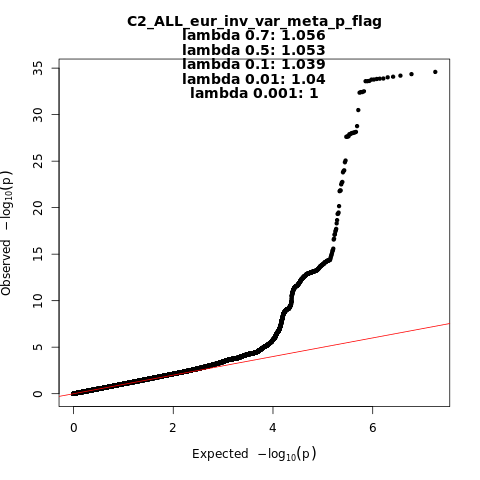
Phenotype
Covid vs. population
Population
Eur
Total Cases
38984
Total Controls
1644784
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 269 | 368 |
| BelCovid_EUR | 485 | 1477 |
| BioVU_EUR | 141 | 70615 |
| CCPM_EUR | 332 | 32375 |
| CU_EUR | 508 | 2149 |
| DECODE_EUR | 4256 | 270934 |
| EstBB_EUR | 1322 | 193774 |
| FinnGen_FIN | 810 | 237901 |
| GCAT_EUR | 253 | 4735 |
| GENCOVID_EUR | 1220 | 2443 |
| GFG_EUR | 147 | 5442 |
| GHS_Freeze_145_EUR | 869 | 112862 |
| Genotek_EUR | 676 | 12317 |
| INTERVAL_EUR | 838 | 40994 |
| LGDB_EUR | 275 | 1313 |
| Lifelines_EUR | 244 | 26553 |
| MSHS_CGI_EUR | 330 | 1396 |
| Stanford_EUR | 169 | 190 |
| TOPMed_CHRIS10K_EUR | 92 | 2373 |
| TOPMed_Gardena_EUR | 452 | 458 |
| UCLA_EUR | 203 | 17391 |
| UKBB_EUR | 6490 | 328577 |
| SPGRX_EUR | 362 | 302 |
| MVP_EUR | 1520 | 7600 |
| genomicsengland100kgp_EUR | 218 | 62302 |
| Helix_EUR | 178 | 5441 |
| MGI_EUR | 122 | 51458 |
| NTR_EUR | 145 | 5252 |
| PHBB_EUR | 151 | 29966 |
| Ancestry_EUR | 2417 | 14933 |
| 23ANDME_EUR | 9913 | 85072 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| HOSTAGE_EUR | 1610 | 2205 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_eur_leave_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_eur_leave_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_C2_ALL_eur_leave_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_C2_ALL_eur_leave_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_eur_leave_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_eur_leave_23andme_20210107.txt.gz_1.0E-5.txt
C2_ALL_eur_leave_ukbb
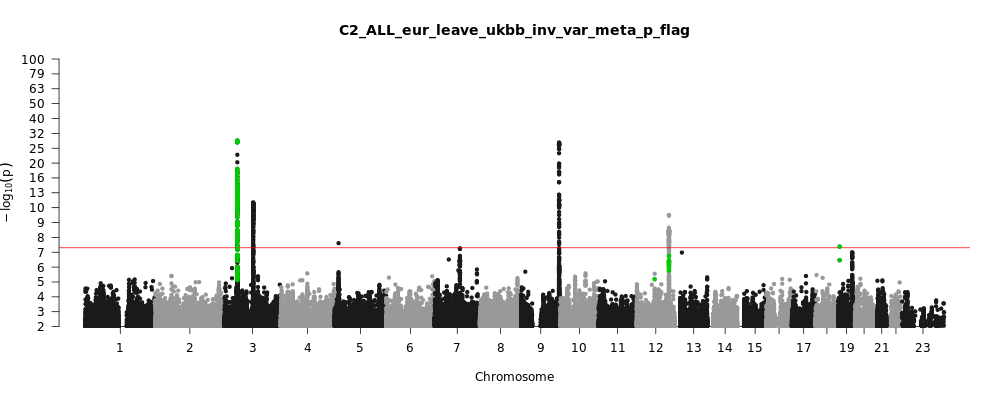
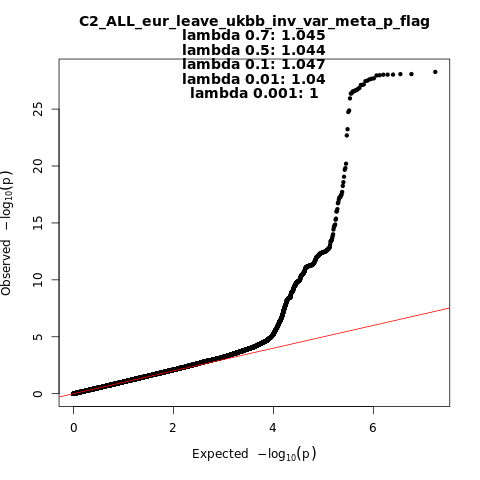
Phenotype
Covid vs. population
Population
Eur
Total Cases
32494
Total Controls
1316207
Contributing Studies
| Name | n_cases | n_controls |
|---|---|---|
| BQC19_EUR | 269 | 368 |
| BelCovid_EUR | 485 | 1477 |
| BioVU_EUR | 141 | 70615 |
| CCPM_EUR | 332 | 32375 |
| CU_EUR | 508 | 2149 |
| DECODE_EUR | 4256 | 270934 |
| EstBB_EUR | 1322 | 193774 |
| FinnGen_FIN | 810 | 237901 |
| GCAT_EUR | 253 | 4735 |
| GENCOVID_EUR | 1220 | 2443 |
| GFG_EUR | 147 | 5442 |
| GHS_Freeze_145_EUR | 869 | 112862 |
| Genotek_EUR | 676 | 12317 |
| INTERVAL_EUR | 838 | 40994 |
| LGDB_EUR | 275 | 1313 |
| Lifelines_EUR | 244 | 26553 |
| MSHS_CGI_EUR | 330 | 1396 |
| Stanford_EUR | 169 | 190 |
| TOPMed_CHRIS10K_EUR | 92 | 2373 |
| TOPMed_Gardena_EUR | 452 | 458 |
| UCLA_EUR | 203 | 17391 |
| SPGRX_EUR | 362 | 302 |
| MVP_EUR | 1520 | 7600 |
| genomicsengland100kgp_EUR | 218 | 62302 |
| Helix_EUR | 178 | 5441 |
| MGI_EUR | 122 | 51458 |
| NTR_EUR | 145 | 5252 |
| PHBB_EUR | 151 | 29966 |
| Ancestry_EUR | 2417 | 14933 |
| 23ANDME_EUR | 9913 | 85072 |
| idipaz24genetics_EUR | 106 | 75 |
| Amsterdam_UMC_COVID_study_group_EUR | 108 | 1413 |
| HOSTAGE_EUR | 1610 | 2205 |
| SweCovid_EUR | 77 | 3748 |
| genomicc_EUR | 1676 | 8380 |
Downloads
GRCh37 liftover: COVID19_HGI_C2_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz
GRCh37 (.tbi): COVID19_HGI_C2_ALL_eur_leave_ukbb_23andme_20210107.b37.txt.gz.tbi
GRCh37 (filtered): COVID19_HGI_C2_ALL_eur_leave_ukbb_23andme_20210107.b37_1.0E-5.txt
GRCh38: COVID19_HGI_C2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz
GRCh38 (.tbi): COVID19_HGI_C2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz.tbi
GRCh38 (filtered): COVID19_HGI_C2_ALL_eur_leave_ukbb_23andme_20210107.txt.gz_1.0E-5.txt
